BLASTing sequences using Case It v6
This tutorial describes two different methods of BLASTing sequences. They both access the BLAST routines at the NCBI web site, but the first method (starting with Step 1 below) does it 'behind the scenes' and returns results as an html file that you open with your web browser. The second method saves the sequence you want to blast as a file and automatically opens your web browser to the NCBI site, so that you can upload and BLAST the sequence.
In addition, you can click on individual fragments on a gel and BLAST them - click here to skip down to this part of the BLAST tutorial.
Return to tutorial menu
Note: BLASTing sequences directly using Case It requires that Java be installed on your computer (if it is not installed, the black window shown in Step 7 below will say that it can't find Java). To install Java for free, go to the Java website of Sun microsystems, and install Java before continuing with this tutorial.
|
1. Begin by using the Clear menu to clear all previous analyses.

2. Use the navigation bar at the bottom of the Data Screen to open the 'Sequence analysis' (SA) window, if it is not already open.

The Sequence Analysis (SA) window appears to the right of the Data Screen.
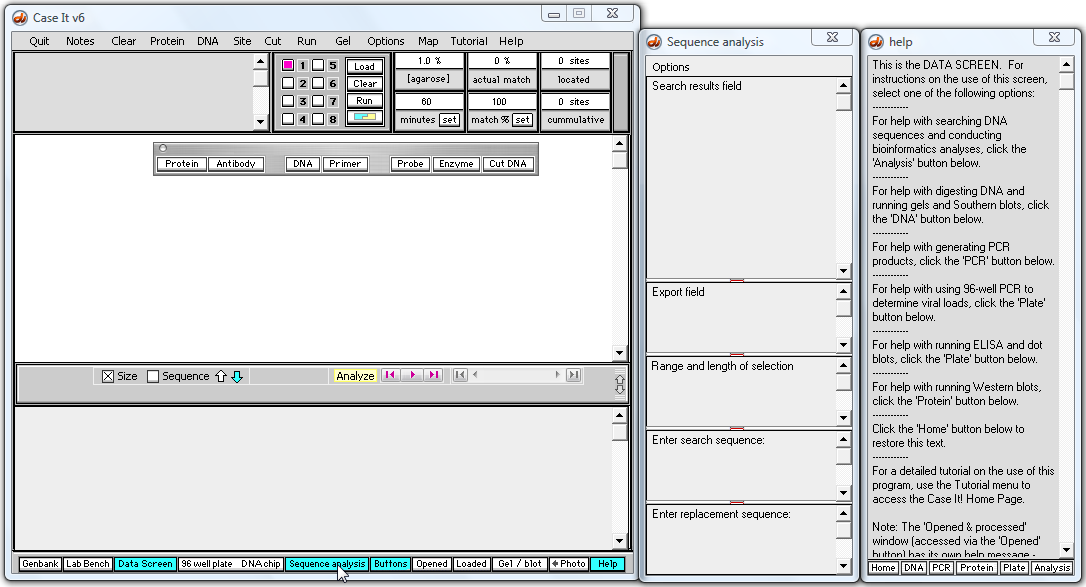
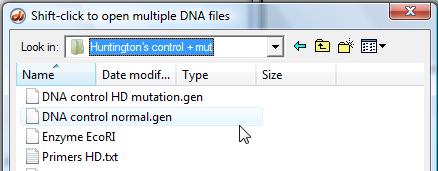
3. Click the DNA button and navigate to the 'Huntington's control + mut' folder, which is inside the 'Tutorials' folder. Open the file 'DNA control normal.gen'.

4. Click the 'Sequence' check box, then highlight the characters shown below (you may have to scroll to the top of the field first). This is the region that contains the triplet 'cag' repeat.
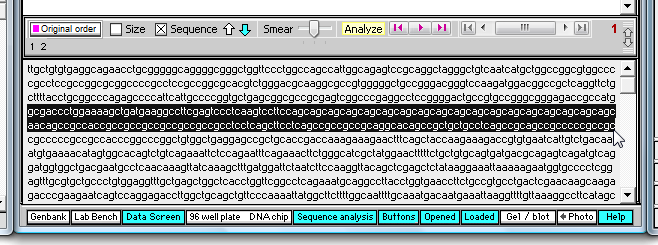
5. Click the yellow 'Analyze' button and make the selections shown below.

6. Note that the highlighted sequence appears in the 'Export field' of the SA window, and that the Export field is automatically resized to make it bigger.
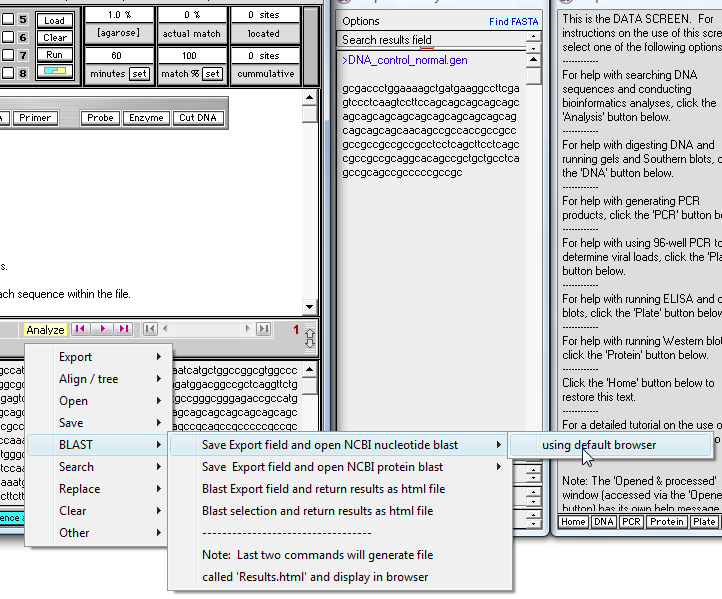
Now us the Analyze menu again and select 'Blast DNA in export field and return results as html file'.
Note: Since the sequence in the Export field is the same as the highlighted sequence at the bottom of the Data Screen, you could also select 'Blast selection and return results as an html file' (the command below the one we have selected) and get the same result, assuming that the sequence is still highlighted.
Note: This first method of blasting only currently works for DNA files. For protein files, you can blast them using the second method described begining in step #11 below. |
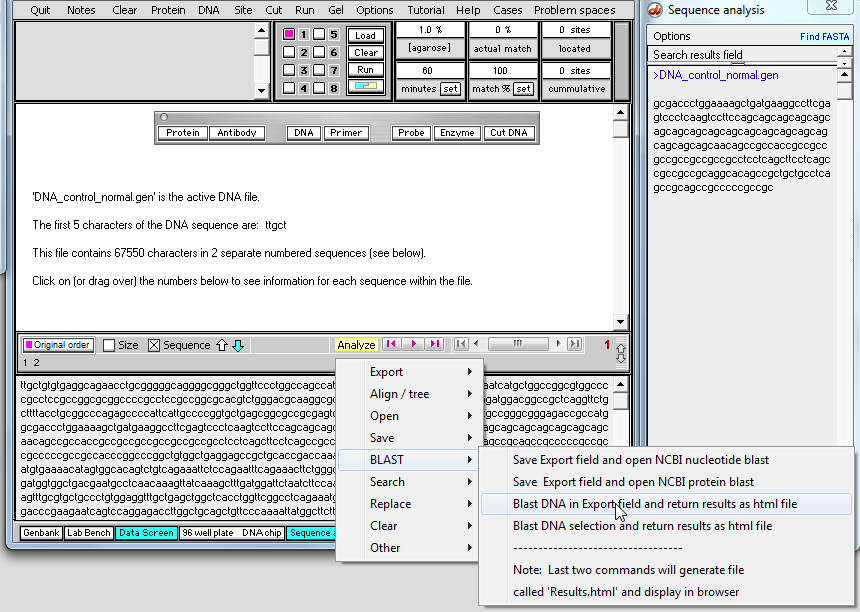
7. The window below automatically appears, showing the sequence that is to be blasted.
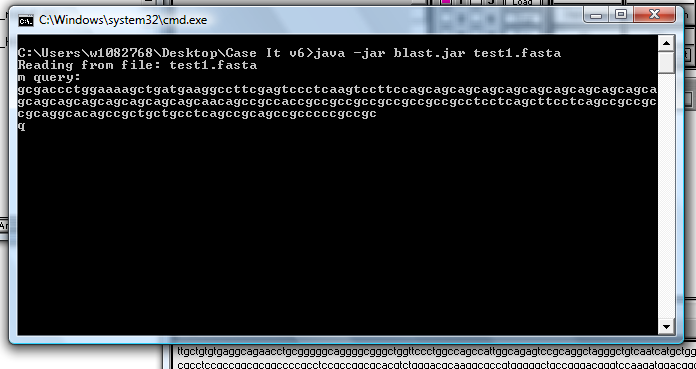
8. After a period of time, lines similar to those below will appear in the window.
| Note: Please be patient - do not do anything until the 'Press any key to continue' line appears. |
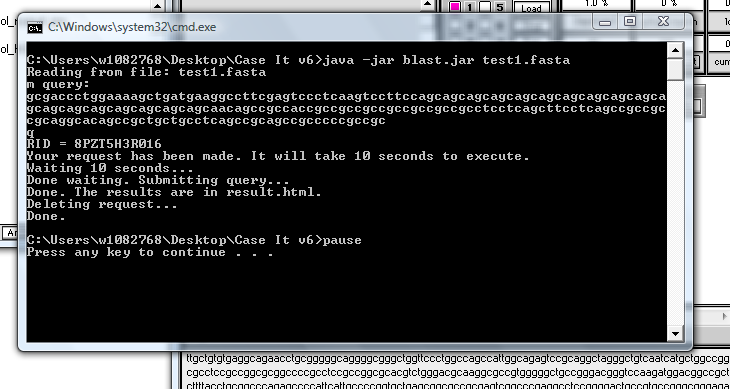
9. Your web browser should open and display the web page automatically, but if it does not, open the Case It v6 folder and double-click on the 'results.html' file to open it in your default web browser.
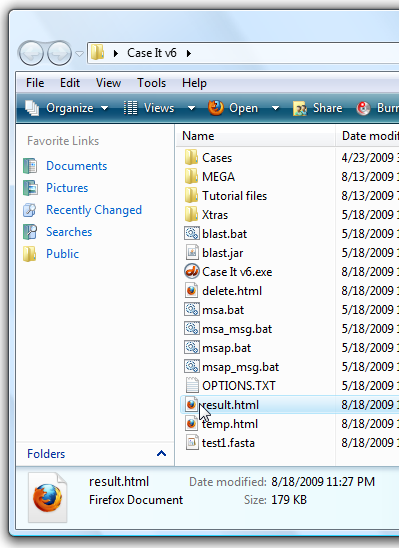
10. Scroll down and find the BLAST results, and note that many of the top hits are identified as the Huntington's disease gene.
| Note: We are in process of changing the formatting of this web page to make it easier to interpret, and will provide additional guidelines for evaluating these results. |
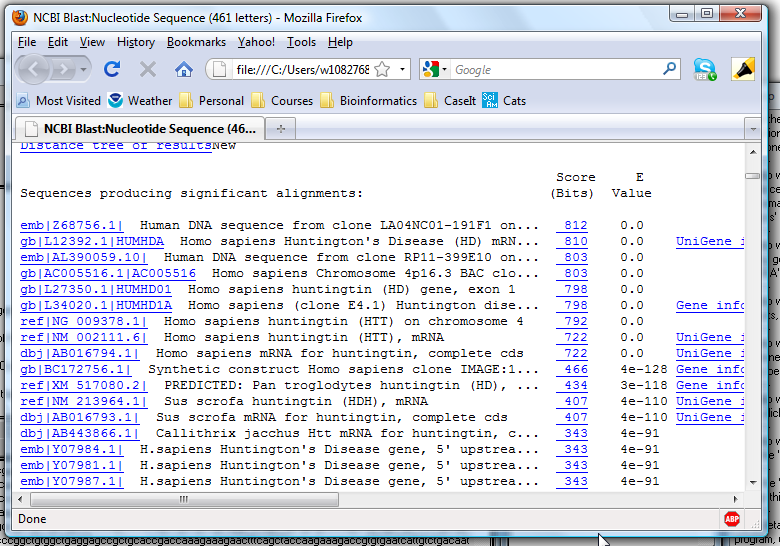
11. The above demonstrated the first method for BLASTing results (doing it 'behind the scenes and returning the results as an html file). We will now demonstrate the second method, which is to save the results of the Export field as a file and open your browser to the NCBI blast page. The previous method is a quicker way to get the BLAST results, but it does not return the complete BLAST page (e.g. graphics of results is missing). In this second method you will go to the actual NCBI site and upload your exported sequence.
If you still have the sequence in the Export window, begin by making the menu selection shown below, via the 'Analyze' button. If not, repeat steps 4 and 5 above first.
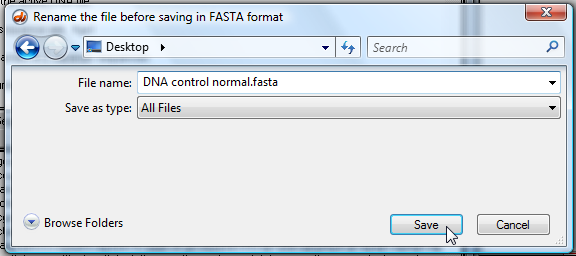
12. You will be asked to rename the file before saving it, so change the default name to one that describes your sequence (a name that you will recognize). Save the file to your desktop to make it easier to find. Note that the file is automatically saved in FASTA format.
| Note: If you do not change the name of the file and attempt to replace an existing file with the same name, it may not replace the file properly. This is a known issue with the software that cannot be fixed. So always rename files instead of replacing existing files when you save them. |
13. After clicking the 'Save' button above, your web browser will start and automatically open to the NCBI nucleotide BLAST page. Click the 'Choose file' button on this page, browse for your saved file.Note that the default setting is to search the 'Human genomic + transcript' database, but this could be changed to search different databases, depending on the nature of your sequence. For this example, the 'Human genomic' database is appropriate. Also, select the 'somewhat similar sequences (blastn)' option towards the bottom of the screen. Finally, click the BLAST button at the bottom of the screen.
| Note: If the URL for this web page changes, you can modify the contents of the OPTIONS.TXT file in the Case It v6 folder, since the URL is read from this text file every time Case It v6.exe opens. You can modify other URL's accessed by the program as well, if they change. |
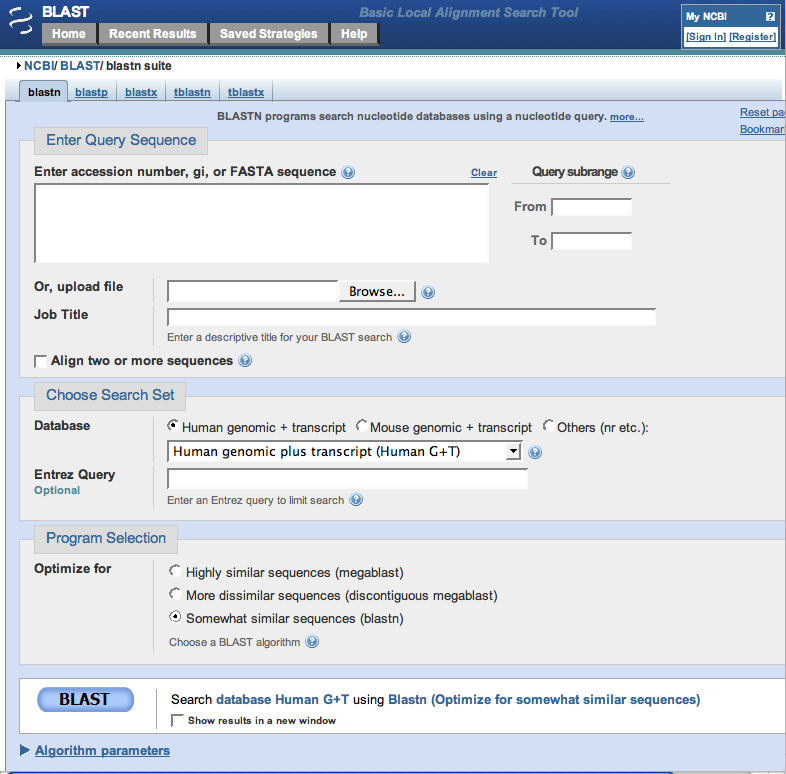
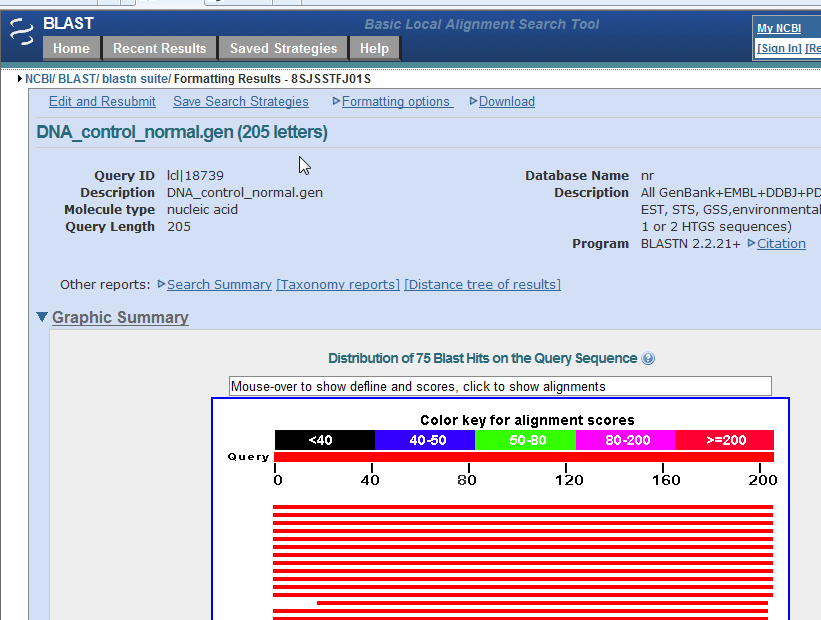
14. Results will appear as shown below (only part of the page is shown in this screen shot). Note that the graphic summary of the results, which was not shown with the first method, is visible.
15. Another option is to click on fragments on a gel and BLAST them, as described below.
Assume that we have digested the mutated and control sequences for the Huntington's mutation, and run a gel (click here for a tutorial on how to do this). We now want to BLAST the fragment containing the repeats to see if other genes have this type of repeat.
Click on the third fragment (or click on the box associated with the third fragment), and note that the size of the fragment appears in the field at the bottom of the Data Screen.
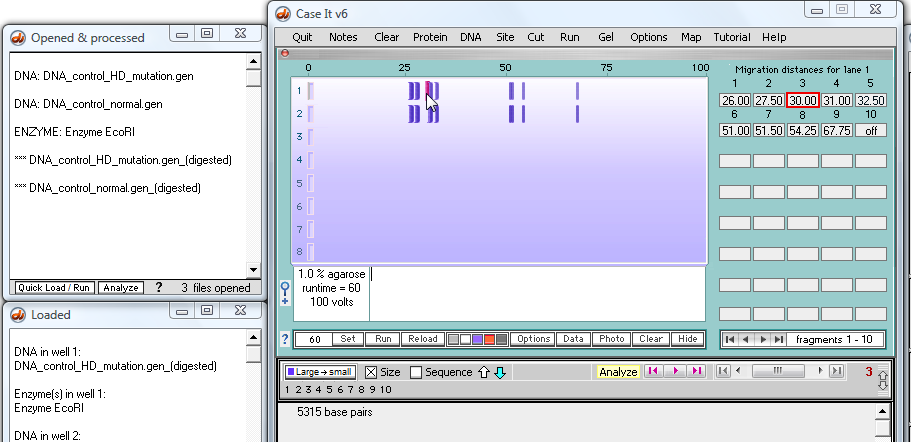
16. Click on the 'Sequence' checkbox to show the sequence rather than the size.
17. Click on the 'Sequence Analysis' button on the navigation bar at the bottom of the screen, then click on the yellow 'Analyze' button and make the menu selection shown below.
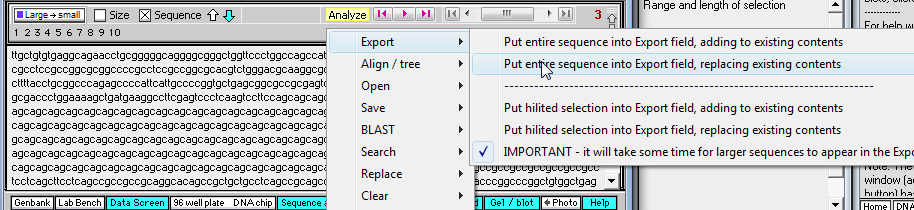
18. The entire fragment sequence is now in the 'Export' field of the Sequence Analysis window.
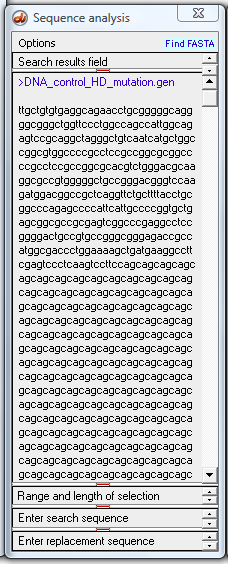
19. Use the 'Analyze' button again and make the menu selection shown below.
| Note: You can also select 'Save Export field and open NCBI nucleotide blast', to give you options of setting parameters via the NCBI web site. If you select this option, you will need to browser for the saved sequence, then click the BLAST button on that web page. |
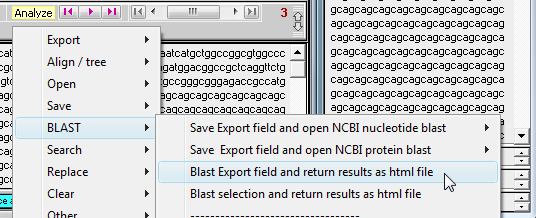
20. The window below appears, indicating that the sequence has been submitted to the NCBI BLAST site. Once the message at the bottom of the screen appears, press any key to continue.
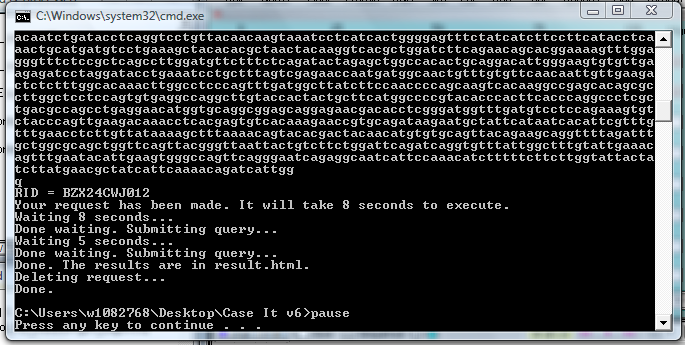
21. Your web browser will open automatically, and display the BLAST results.
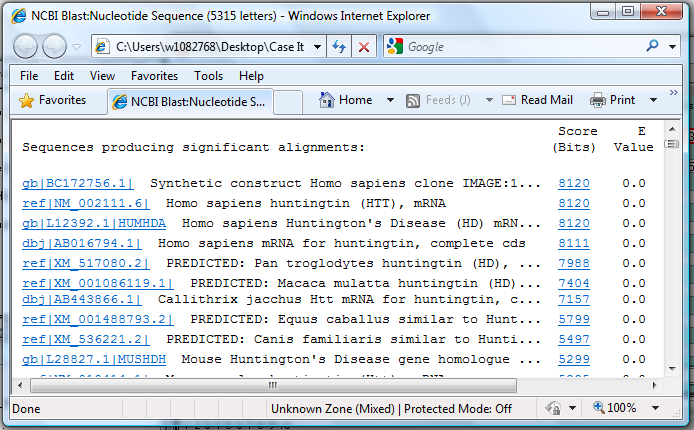
You could also use the other method of blasting, which is to save the contents of the Export field and open your web browser to the NCBI site. This method is described starting in Step 11 above.
This concludes this tutorial.
Back to tutorial menu






















