Answers to questions in Part 2
5. Why should this undigested DNA sequence be loaded into a well?
The undigested sequence from chromosome 18 serves as a control, for comparison against the digested sequence to be loaded into the second well.
20. Why was the DNA sequence cut at this particular location? What three other ‘standard’ PAM sequences could have enabled Cas9 to cut at this location?
The sequence was cut 3 bp ‘upstream’ of the PAM sequence (GGG). The other three possible PAM sequences are AGG, CGG, or TGG, since the ‘standard’ PAM for Cas9 is 5′-NGG-3′.
24. Consider the amount of information in a word processor document with 19,600 pages. Chromosome 18 is one of the smaller chromosomes, containing only about 2.6 percent of the total information in the human genome. How many pages would it take to write out the entire human genome?
(19,600 x 100)/2.6 = 753,846 word processor pages
32. The sequence above would actually be on the ‘non-target’ strand of the DNA molecule, because that is what the Case It simulation will use to find the location on the chromosome 18 sequence (recall from Part 1 that the PAM, if present, must be on the ‘non-target’ strand). In reality, how does the CRISPR-Cas9 recognize the location, and how is it involved in cutting it?
For this example, the guide RNA sequences comes into place along the target strand of DNA by complementary base-pairing, as shown in Part 1. Guide RNA has a ‘handle’ (scaffold) that is used to bring Cas9 into position to do the actual cutting. More detail can be found here.
35. Using this number as a reference, determine the exact location of the 5′-NGG-3′ PAM site.
The cut is three bases from the end of the recognition site for the guide RNA used, at location 31,593,004 (AAAGGCTGCTGATGACA |CCT). This tells you that the 5′-NGG-3′ PAM site must be on positions 31,593,008, 31,593,009, 31,593,010 of the chromosome 18 sequence (AAAGGCTGCTGATGACA |CCTNGG).
44. What is an exon? Why was it so important that the cut occur in an exon of the TTR gene, rather than in an intron?
Exons are the parts of a gene that are ultimately ‘expressed’ as protein, where as introns are the parts that are removed before mRNA is spliced together. So to ‘knock out’ the TTR gene the cut must interfere with the sequence of DNA in an exon.
Answers to questions in Part 3
13. There are four mismatches within 12 bases of the PAM. Look again at information on guide RNA design in Part 1 (then hit the back arrow of your browser to return here). Based on this information, how likely is it that Cas9 would make an off-target cut at this particular location?
Since there are four mismatches within the 12 bases next to the GGG PAM location, it is highly unlikely that Cas9 would cut at this location (the 70% match setting of Case It allows for mismatches anywhere along a sequence). Also, the GC content of the 70% match is only 40%, which is at the low end of the acceptable range.
21. Note that the off-target effect is not found directly in a gene (designated here by colored lines). Does this mean that an off-target cut here will have no harmful effect?
Not necessarily. An unidentified gene could be present at that location, or it could contain information necessary for the operation of adjacent genes. But again, it is highly unlikely that a cut would be made at that location based on the answer to question 13 above.
39. Will Cas9 cut chromosome 1 at this location? What are the factors that determine whether Cas9 will cut DNA at a particular location? If it did cut here, would it necessarily ‘knock out’ this gene? Note that the gene is named C1orf127. Here are links to the gene in Wikipedia and also in the NCBI Gene database. What would be the effect of knocking out this particular gene?
Although there is an 85% match against the 20 bp guide RNA sequence, with a 100% identity (17/17 matched exactly), Cas9 would not cut at this location because there is no NGG PAM sequence at the 3′ end of the sequence. So the location would definitely not be cut by Cas9. But even if a PAM sequence had been present, the cut would have occurred in an intron, which is not expressed by the gene. According to the references, this gene has unknown function but is secretory in nature.
44. Does this result necessarily mean that Gillmore’s team used the ‘wrong’ guide RNA to knock out the TTR gene? What are some of the reasons why Gillmore’s team used a guide RNA sequence different from the ones that Synthego’s tool recommended?
No, it does not mean that they used the ‘wrong’ guide RNA. According to Synthego, their recommendations are based on information only in publicly available databases, and Gillmore’s team used a complex protocol that included in vivo and in vitro tests for off-target effects. The guide RNA they did use was 11th on the list generated by Synthego’s tool.
Answers to questions in Part 4:
24. The guide RNA/Cas9 complex that Gillmore’s team used cut the gene in exon 2. Would a mutation that is located on exon 3 be disabled by a cut in exon 2?
The goal is to disable the gene, and a cut in exon 2 would most likely do this. Cuts are repaired in such as way that the rejoined DNA can no longer be part of a functional gene.
Answers to questions in Part 5
1. The use of NHP (non-human primates) in research has been controversial. This article proposes an ethical framework for such studies. What is your personal opinion regarding the ethics of such testing?
The linked article would generate a good discussion of this important topic.
5. From Part 2, you know that the human chromosome 18 sequence has a size of 80,373,285 bp. What are some reasons for this apparent difference in size between human and monkey chromosomes?
Although evolutionary divergence is responsible for these differences, it is striking how many genes humans and macaques have in common. A good student exercise would be to browse the genomes in the vicinity of the TTR gene to see these similarities.
17. Is this the same exon targeted by the guide RNA used for the human trials, as determined in Part 2? How might this affect interpretation of results of monkey vs. human trials?
The monkey guide RNA target site is on exon 1, and the human target site is on exon 2. Using different exons might influence off-target effects of the drug regimen for monkeys and humans, although the authors say they did extensive analysis for these effects.
18. The guide RNA sequence target sequence used for the monkey trial (ACACAAATACCAGTCCAGCG) was different than the one used for the trial on human patients (AAAGGCTGCTGATGACACCT). Why did the investigators use different guide RNA sequences for the two trials?
Different exons were targeted for monkeys vs. humans, meaning that the guide RNA target sites had to be different (see below).
23. The ‘Indel Frequency’ on the right of Fig. 3B shows that the vast majority (98.9%) of the indels were single base-pair insertions (A). 1.03% were insertions with more than one base, designated by the letter N. These insertions were either AA or AGG. Would either of these have resulted in a stop codon in exon 1 of the monkey TTR gene?
The only indel that causes an obvious stop codon in exon 1 is the single base-pair insertion, as shown in Step 23 of Part 5. The authors do not specify the specific frame shift mutations causing gene knockout, for either the human or monkey trials.
24.
(1) Assuming that the most common indel is a one bp insertion or deletion, can you find a stop codon in exon 2 of the human TTR gene? Remember that if one can be found, it must be downstream (toward the 3′ end) of the DNA sequence. The three DNA stop codons in humans are TGA, TAA, and TAG. Can you find a stop codon if the mutation had more than one insertion or deletion? Recall that in the monkey trials, 99% of the idels were a single base pair insertion.
No stop codon resulting from a single base pair insertion or deletion is obvious downstream of the indel site in exon 2. The authors state that next-generation sequencing data show that that frameshift mutations were responsible for gene knockout in humans, but do not provide supporting data in the paper or in its supplement.
(2) Is there a potential stop codon in exon 3? Considering the mechanism for splicing RNA after transcription but before translation, is it physically possible for a frameshift mutation in exon 3 to be induced by a frameshift mutation in exon 2?
If it was physically possible for a frameshift mutation in exon 2 to cause a stop codon in exon 3, then such a stop codon can be found. However, based on the mechanism for gene splicing, it appears that this would not occur because a frameshift mutation in one exon would not affect the way that the beginning of an exon is recognized in the exon downstream from it. The authors make no statement to this effect, one way or the other.
(3) In the legend to Figure 2 of Gillmore et al. 2021, the authors say that the “primary indel patterns were a single-nucleotide deletion or insertion at the cut site, inducing a frameshift mutation (data not shown).” They are referring to human hepatocytes (liver cells) studied in vitro. Why did they not show the data for the human trials, when they did show the data for the monkey trials in Fig. 3B?
It is unclear why the authors did not show this important data, especially since the single-nucleotide cut site for the monkey trials is shown, in Fig. 3B. One possibility is that the private company that sponsored the research considered the information proprietary. Another possibility is that the editor of the article was saving space, but that seems unlikely considering the critical nature of the data.
Note: The sequence in the file DNA TTR gene monkey.txt is the reverse complement of the original Macaca fascicularis TTR gene sequence obtained from Genbank. This was necessary so that Case It could search the coding strand for both the monkey and human TTR genes. The human TTR gene was originally entered into Genbank as the coding strand, but the Macaca fascicularis gene was entered as the template strand. Unlike the Genome Data Viewer, Case It can only search one strand of a DNA sequence, so the coding strand was the one selected for this purpose. By taking the reverse complement of the monkey sequence, it was ‘converted’ from the template strand to the coding strand.
41.
(1) For both exon2 and exon1, the mismatch is located relatively close to the PAM (4 bp away for exon 2, and right next to the PAM for exon 1). Is it possible for Cas9 to cut when mismatches are located this close to the PAM?
It is unlikely that Cas9 could cut with mismatches so close to the PAM.
(2) How might Gillmore’s team have dealt with this, if they wanted to use the similar guide RNAs for both the human and primate trials?
They could have modified the monkey guide RNA by changing a single base, so that it matched the corresponding site in exon 1 of the human TTR gene. There is no evidence from the published article that the authors tried this approach, but they might have done so.
(3) Which would give better results, using different guide RNAs that target different exons with a 100% match, or using 95% similar guide RNAs that target the same exon?
The authors conducted extensive studies using a variety of procedures to minimize off-target effects of the treatment, but they do not address this question so there is no way of knowing if this approach was considered.
(4) Here is a link to a statement from Gillmore’s protocols giving the reason for their decision to use separate guide RNAs for the human and monkey trials, and another link describing why crab-eating macaques were used as primate surrogates. Is there any conflict between these two statements?
In the protocol for the study, the authors state that “due to the highly species-specific activity of NTLA-2001 to its human genomic target, a cynomolgus surrogate LNP [lipid nanoparticle]…was developed to enable pharmacologically relevant assessments in nonclinical studies.”.
But they also state that “the NHP [non-human primate] species is closely related, both phylogenetically and physiologically, to humans and is an acceptable non-rodent species for nonclinical PK, PD, and safety evaluations of gene therapies.” More information from the authors would be required to resolve this apparent conflict between the two statements.
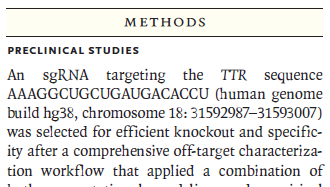
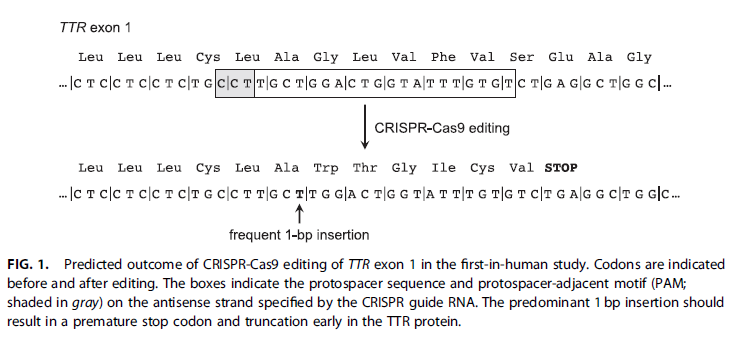
(5) The quote and figure below are from a news article in the CRISPR Journal about Gillmore’s study (“CRISPR Hits Home in a First-in-Human study”,Musunuru 2021). The author of this article had not read Gillmore et al. 2021 carefully, and made a major assumption that was in error. What was that assumption? Hint: Compare Fig 1. below with the data in Step 41 above, and also with the diagram in Step 23.
The author of the news article missed the first sentence of the Methods section of Gillmore et al. 2021 (see image below), and then assumed that they targeted exon 1 in both the monkey and human trials, when in fact the guide RNA used for the human trials targeted exon 2.
The author assumed that Gillmore’s team must have modified the monkey guide RNA so that it would target the human exon 1 site, because the human target site is what is reported in Figure 1 of the news article. The first three letters in the next two lines are the PAM site, shown here underlined, with the base change in red. That letter is the first base in the 20 base target sequence, in boldface. See the answer to question 41.(2) above.
Monkey site on exon 1
CTCCTCCTCTGCCTCGCTGGACTGGTATTTGTGTCTGAGGCTGGC
Human site on exon 1 – same as in the figure below
CTCCTCCTCTGCCTTGCTGGACTGGTATTTGTGTCTGAGGCTGGC